
Unit B: Coupled biocatalytic reactions
Our Challenge
We aim at understanding natural catalysts, namely enzymes. Enzymes in cells are able to perform complex coupled catalytic processes. These function effectively, maybe more efficiently than in artificial human-engineered catalysts. Biocatalytic processes ‘only’ need to be deciphered or recombined in alternative ways. This is our challenge!
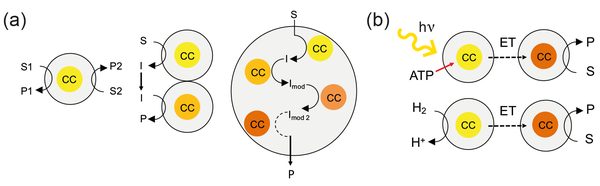
With this knowledge, we'd like to design smart catalysts. For example, we wish to engineer electron transfer chains.
Our approach
Our efforts are first directed to understanding in detail natural enzymes. We focus on the catalytic mechanisms and dynamics of biocatalysts capable of controlling coupled biocatalytic reactions. Our key questions are:
- How are two catalytic reactions coordinated at a single active site?
- How does the interaction between active sites work in enzymes? Here, for instance, we are interested in substrate channeling, sequestering of reactive intermediates and servicing between active sites.
- How is the electron transfer between enzymes controlled?
- How can we design catalytic systems learning from nature?
Our studies strive to elucidate the molecular bases of various aspects of catalytic multi-functionality.
A particularly challenging example are bi- and multifunctional carbon monoxide dehydrogenases, in which the catalytic activities of three coupled metal-containing active sites are coordinated in a way that allows the efficient conversion of CO2, CoA, and CH3 + into acetyl-CoA. We will analyze how these centers communicate and their different oxidation/catalytic states trigger conformational changes in order to sequester and channel the one-carbon intermediates.
Keeping electrons at a high energy level between separate oxidative and reductive reactions is a widely employed concept in nature to enable biocatalytic processes. This concept will be explored for O2-tolerant hydrogenases and formate dehydrogenase, and subsequently exploited for coupling these biocatalysts with electron-demanding processes such as CO2 reduction.
Team
Robert Bittl
(Pulsed) EPR spectroscopy
Holger Dau
X-ray spectroscopy, QCL-IR/FTIR
Holger Dobbek
Enzymology, X-ray crystallography, Fe/S-enzymes
Matthias Driess
Molecular (heterobimetallic) active site mimics
Matthias Gimpel
Gene regulation, Bioprocess development
Peter Hildebrandt
Raman spectroscopy (surface-sensitive, in situ)
Marius Horch
Ultrafast, multidimensional and in vivo vibrational spectroscopy
Oliver Lenz
Biochemistry, enzyme engineering, hydrogenases
Christian Limberg
Model compounds, O2- and CO2-activation
Maria Andrea Mroginski
Theory (QM/MM, MD)
Henrike Müller-Werkmeister
(transient) 2D-IR & ultrafast UV-Vis spectroscopy
Peter Neubauer
Bioprocess engineering, enzyme overproduction
Ariane Nunes Alves
Molecular modeling and simulation of ligand-protein interactions
Kallol Ray
Bioinorganic chemistry, O2 activation
Patrick Scheerer
X-ray crystallography, XFEL
Petra Wendler
Cryo electron microscopy
Ingo Zebger
IR spectroscopy (surface-sensitive, in situ)
Athina Zouni
Biochemistry, X-ray crystallography, XFEL





















