With the development of ever more powerful computer systems and new calculation methods, the importance of computer-aided predictions has also increased in many areas of research and development. For example, predicting the kinetics of drug-protein binding based on calculations has become increasingly important for the pharmaceutical industry. Thus, computer-based predictions now support experimental studies in the development of new drugs.
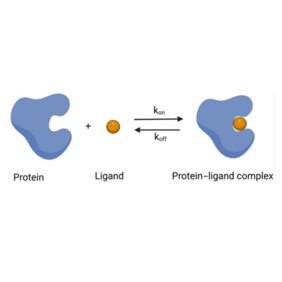
Understanding the binding mechanism of drugs to their target proteins can be helpful in drug development, especially in predicting drug efficacy. The binding mechanism is described by kinetic binding rates, which can be predicted computationally. This has led to a growing interest in developing computational methods to gain insight into binding kinetics and mechanisms, and to date many methods have been developed for this purpose.
In their current review, the UniSysCat researchers Farzin Sohraby and Ariane Nunes Alves summarize and compare computational methods to predict ligand binding kinetics that were published from 2019 onward and applied to two systems, trypsin–benzamidine and kinase–inhibitor complexes. They chose these two model systems due to their benchmarking character and the availability of many publications, which facilitates comparison across different methods.
The reviewed computational methods can not only be used for prospective predictions of kinetic rates but also to obtain mechanistic insights into selectivity and drug resistance. In the last years, machine learning methods greatly benefited from the availability of large and consistent datasets, obtained under similar experimental conditions. Yet, the authors note that there is still a great need for experimental data for benchmarking of computational methods.
Finally, the authors suggest different ideas to improve the predictions of kinetic rates with high errors, such as the use of scaling factors for methods which change the potential energy surface, careful parametrization of ligands, and extensive sampling of bound, transition, and unbound states. They furthermore suggest to combine quantum mechanics with enhanced sampling methods to study protein–ligand binding in order to reduce errors.
This review has been published in Trends in Biochemical Sciences: F. Sohraby, A. Nunes-Alves, Advances in computational methods for ligand binding kinetics, Trends in Biochemical Sciences (2022). https://doi.org/10.1016/j.tibs.2022.11.003