A Comprehensive Discovery Platform for Organophosphorus Ligands for Catalysis
Tobias Gensch, Gabriel dos Passos Gomes, Pascal Friederich, Ellyn Peters, Théophile Gaudin, Robert Pollice, Kjell Jorner, Akshat Kumar Nigam, Michael Lindner-D'Addario, Matthew S. Sigman, Alán Aspuru-Guzik
The design of new catalysts can be tedious and is sometimes not very successful: it is often dependent on the intuition of the respective scientists and limited to local structural searches. In order to simplify and accelerate the creation of new catalysts, it is advantageous to be able to predict the performance of novel catalysts and generate a limiting template in the space of infinite possibilities. Data-driven workflows embedded in a library of potential catalysts can be used to build predictive models for catalyst performance and serve as a blueprint for novel catalyst designs.
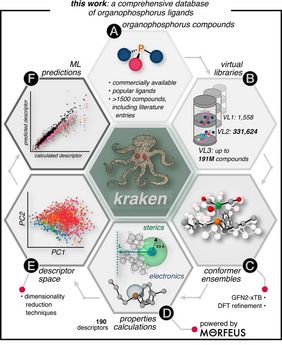
In this context, UniSysCat group leader Tobias Gensch as part of an international team of researchers introduces kraken: „an extensive virtual open-access library covering monodentate organophosphorus(III) ligands targeted at facilitating the design and optimization of catalytic processes.“
The kraken library has been build in several steps. The researchers first collected 1558 organophosphorus compounds from literature and commercial sources as a basis for the library. In the next step, conformer ensembles were generated at a semiempirical level (GFN2-xTB) of theory and reoptimized and analyzed by DFT. For all 21437 conformers that were evaluated employing this procedure, 190 different physical-organic properties and other representative descriptors were then stored and with these, chemical property spaces could be determined. With this large underlying data set, machine learning models could be trained to predict the properties of compounds at an even larger scale of the library consisting of 331776 compounds with up to two different substituents per ligand or even over 191 million entries when all three substituents can be different. Thus, the library created covers a vast number of possible organophosphorus(III) ligands for catalyst design.
Finally, the kraken library is intended to promote inverse catalyst design: "We envision that it will enable synthetic chemists to perform computer-assisted interactive ligand exploration and provide new insights into relevant properties to solve a given problem. The kraken tool may enable informed catalyst design based on organophosphorus ligands, facilitate the optimization of reaction process parameters, inspire new ligand choices, and promote the synthesis of new organophosphorus compounds."
Is the database intended only for theoreticians or also for experimental chemists?
Tobias Gensch says, they hope that both, theoretical and experimental chemists will benefit from kraken. One by using data-driven techniques to gain more information from existing experiments and by saving time and ressources. The other by applying and further developing the database to ultimately develop even better tools.
How exactly can Kraken be used for catalyst design? The chemist in the lab enters a few properties and the database spits out suggestions for catalyst candidates?
Unfortunately, it is not quite that simple yet. The database provides descriptors that can be used to develop statistical models linking experimentally observed reactivities (i.e. yield or selectivity etc.) to the structure of the catalysts. These models can then be applied to the remaining ligands in "kraken" in order to predict their reactivity and thus obtain suggestions for new, better catalysts. In the last part of the article, we demonstrate this process with an example from the literature.
However, the process is still so complicated that it is better if it is accompanied by an experienced theoreticians to identify problems and get better predictions. Yet, to give an outlook: My goal - and that of many colleagues - is to develop tools that are as simple as possible to use and can be routinely applied in pure experimental groups.
What exactly can be downloaded from the database?
We provide the underlying properties of the individual conformers as well as their structures (as xyz-file) and in principle also the output files of the XTB and DFT calculations.
Kraken is now accessible as a web application (https://kraken.cs.toronto.edu). The open-source nature of the codes and databases is designed to be extended by the community. The findings of Gensch et al. have been published in the Journal of the American Chemical Society: Tobias Gensch, Gabriel dos Passos Gomes, Pascal Friederich, Ellyn Peters, Théophile Gaudin, Robert Pollice, Kjell Jorner, AkshatKumar Nigam, Michael Lindner-D'Addario, Matthew S. Sigman, Alán Aspuru-Guzik A Comprehensive Discovery Platform for Organophosphorus Ligands for Catalysis. J. Am. Chem. Soc. , 2022, XXXX,XXX,XXX-XXX, https://doi.org/10.1021/jacs.1c09718