Protein structure prediction with in-cell photo-crosslinking mass spectrometry and deep learning
Kolja Stahl, Andrea Graziadei, Therese Dau, Oliver Brock and Juri Rappsilber Nature Biotechnology (2023) https://doi.org/10.1038/s41587-023-01704-z
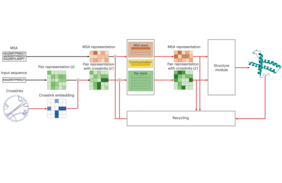
In a joined effort the Rappsilber and the Brock group stemming from the two clusters of excellence UniSysCat (Unifying Systems in Catalysis) and SCIoI (Science of Intelligence), respectively, developed a new prediction tool, AlphaLink, for challenging protein structure targets by combining experimental data and deep learning.
AlphaLink is a modified version of the famous AlphaFold2 algorithm, which gained substantial attention in the scientific community over the last years, as it capably predicts a protein's 3D structure from its amino acid sequence based on an Artificial Intelligence (AI) system, within accuracies that are competitive with experiments. However, challenges remained for proteins undergoing conformational changes or for which only a few homologous sequences are known. AlphaLink incorporates now experimental distance restraint information into the network architecture. It utilizes sparse experimental contacts as anchor points for achieving a higher prediction accuracy. This was experimentally proven by the Rappsilber group which employed a novel approach to obtain residue–residue contacts inside cells by pairing crosslinking mass spectrometry with photo-crosslinking through incorporating into the cellular proteins the noncanonical amino acid photo-leucine. The newly developed tool predicts distinct conformations of proteins on the basis of the provided in-cell distance restraints. This is a move of structural biology into cells and demonstrates the value of experimental data in driving protein structure prediction by a deep learning approach developed by the team. In such way, the noise-tolerant framework for integrating data in protein structure prediction opens a new path for characterizing protein structures accurately from in-cell data.