The study of the structure and function of proteins, which are important building blocks of all living organisms, is quite complicated because the fragile giant molecules often lose their properties when removed from their native environment. A team led by UniSysCat group leader Prof. Dr. Adam Lange from FMP Berlin recently presented a method with which they can extract membrane proteins while preserving their natural environment and investigate the influence of the environment on the activity of the protein.
About a third of all human proteins are membrane proteins, and these are targets for more than half of all drugs, which makes them very important in medicine. However, determining the structure and function of membrane proteins remains a particular challenge for experimentalists, as it is extremely difficult to maintain the native conformation of the protein in isolation from the membrane. The structure and activity of these proteins is however very strongly dependent on the interaction with the lipids that surround them in the membrane.
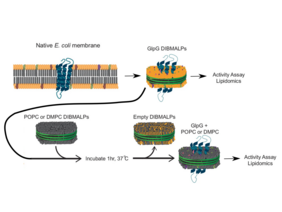
The idea is therefore not to study membrane proteins in isolation, but in their environment, together with the important lipids. This is why nowadays it is not individual membrane proteins that are extracted, but a section of the membrane in so-called nanodiscs. These are synthetic model membrane systems which assists in the study of the proteins. One possibility to mimic the native lipid membrane in such a nanodisc is by using synthetic polymers, for example Diisobutylene-maleic acid lipid polymers (DIBMALPs).
In their recent study, the researchers around Adam Lange used DIBMALPs to extract and study the model membrane protein „E. coli rhomboid protease GlpG“. The team was not only able to extract the protein in a DIBMALP maintaining its function, they went one step further: They used a new experimental technique called „collisional lipid mixing“ to exchange the native lipids in the DIBMALPs with other lipids. Thus, they could directly investigate the influence of different lipids on the membrane protein. They could show, that this mixing of lipids had no deleterious effects on the stability or structure of the studied protein.
So far, the method of collisional lipid mixing had only been applied to DIBMALPs containing no proteins but only lipids. The current study now shows that lipid exchange is not limited to pure lipid-polymer nanodiscs, but also takes place in protein-containing DIBMALPs. The study therefore opens up new possibilities: The method can be used to observe the interactions of proteins and lipids in the membrane and to measure the activity of membrane proteins.
This study has been published in Nature Communications: Sawczyc, H., Tatsuta, T., Öster, C. et al. Lipid-polymer nanoparticles to probe the native-like environment of intramembrane rhomboid protease GlpG and its activity. Nat Commun 15, 7533 (2024). https://doi.org/10.1038/s41467-024-51989-0