Natural protein can be used as scaffolds to engineer enzymes with new substrate specificities. However, new functionalities are commonly associated with significantly lower activity compared to the natural one - so a new function comes at the price of low efficiency. For natural catalytic reactions, nature had billions of years to evolve optimized enzymes tuned to perform their individual tasks. For man-made enzymes, where specifically designed biocatalysts are highly desired, the question thus arises how to achieve this goal within laboratory timescales.
A fascinating approach to novel proteins is changing the building blocks (the 20 canonical amino acids) as part of the genetic code. In form of noncanonical amino acids, this allows equipping proteins and peptides with new functionalities and chemical handles. More than 200 in number, their ribosomal incorporation via expansion of the genetic code opens the way for targeted design of biomolecules. For such new-to-nature amino acids, the machines involved in protein synthesis themselves must be engineered – the aminoacyl-tRNA synthetases. Again, the generation of efficient enzyme variants for new amino acid substrates is a tough challenge, given by the vast sequence space. To engineer new substrate specificity, even randomizing only active residues (e.g. 30 in number, which are structurally close to the substrate) affords a combinatorial space of 2030 variants, which would need to be tested for the desired function. Experimentally, however, only 5-7 positions can be randomized on genetic level and subjected to testing.
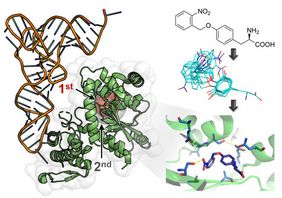
Nediljko Budisa and his team engineer aminoacyl-tRNA synthetases (aaRSs) to provide access to the ribosomal incorporation of noncanonical amino acids. To engineer efficient aaRS variants and to overcome the limitations for screening targeted mutagenesis libraries, computational enzyme design was applied. This allowed generation of a focused aaRS gene library which can be experimentally handled. In total, 17 first- and also second-shell residues, which are more distant to the active site, were diversified in the process. Subsequent cycles of positive and negative aaRS selection resulted in a high-activity enzyme variant for photocaged ortho-nitrobenzyl tyrosine (ONBY), the synthetic aaRS substrate chosen for this study. The obtained aaRS carries ten mutations and outperforms previously reported ONBY-specific enzymes isolated from traditional libraries. The efficiency of amino acid incorporation in living bacterial cells matched that of the wild-type enzyme, the template biocatalyst acylating the tRNA with tyrosine. This now allows the efficient production of ONBY-labelled peptides and proteins, which carry the synthetic photocaged amino acid.
“These results exemplify how computational protein redesign can facilitate the generation of efficient biocatalysts and may lead to an improved general strategy for aaRS library design and engineering.”, says Tobias Baumann, lead author of the current work.
https://www.mdpi.com/1422-0067/20/9/2343